| |
The main focus of our research group is structural proteomics using mass spectrometry (MS), with a focus on chemical cross-linking methods. With the acquisition of a Synapt HDMS (Waters), ion mobility spectrometry has alsobeen recently implemented in our lab. To improve studies from the data acquired using these techniques, molecular modeling in also used (see the instruments available in our lab under “Instruments”). Below is a brief description of cross-linking and ion mobility and what we are developing in each field.
Chemical Cross-Linking

Chemical cross-linking is the process of covalently joining two molecules by the use of cross-linking agents. The molecules to be linked may be proteins, peptides, drugs, nucleic acids or even solid particles. Based on the cross-linker spacer arm length, distances constraints obtained are used to model the structure of the protein and to reveal the interacting regions in the case of protein complexes [Back et al; J. Mol. Biol. 2003, 331, 303–313]. Our research group has been employing chemical cross-linking to study proteins and peptides and, despite the ever increasing number of studies using cross-linking coupled to MS, there are few works that report how the cross-linked peptides fragment or how the presence of the cross-linkers influences the cross-linked peptide dissociation. Based on this fact, fundamental studies on fragmentation of cross-linked peptide species have been done in our lab [Iglesias et al; J. Am. Soc. Mass Spectrom. 2009, 20, 557–566]. We are also testing the reactivity of different residues when reacted with commercial cross-linkers. The knowledge acquired in theses studies has been applied in the identification of cross-linked peptides from protein experiments.
Ion Mobility Mass Spectrometry
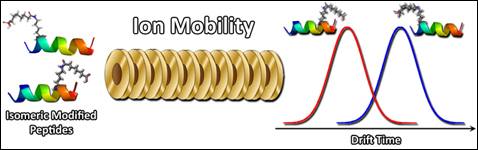
Ion Mobility Spectrometry (IMS) relies on separation of a packet of ions based on mobility differences as they drift through an inert gas under the influence of a weak electric field, which accelerates highly charged ions to a greater extent in comparison with low charge states. Ions with larger collision cross-sections suffer more collisions with the gas and thus have longer drift times, whereas small ions collide less and have shorter drift times. Mobility separation then occurs based on differences of charge state and cross section of the studied ions. An estimation of an ion’s collision cross section by means of IMS experiments offers information about its shape or conformation. We are now working on optimizing instrumental parameters for IMS experiments of model unmodified and modified proteins and peptides. |
|